Coarse Grain Models for Molecular Dynamics Simulations
Computational resources continue to rapidly increase allowing theoretical investigation of larger systems on longer timescales. Indeed the capacity of all-atom (AA) molecular dynamics (MD) has reached a level that permits somewhat routine exploration of systems containing on the order of hundreds of thousands of atoms for timescales approaching hundreds of nanoseconds. Nonetheless, with these spatial and temporal scales, many soft matter and biological systems of interest still extend far beyond this capability.


In order to overcome this issue, techniques such as enhanced sampling methods or reduced description models can be used, just to name a couple. Models using a reduced description of the system are typically referred to as coarse grain (CG) models. Recently there has been a renewed interest in CG techniques with numerous models now appearing in the literature. As well, there are numerous methods for deriving parameters for CG models.
However, each method has its limitations, which has limited greater adoption of these methods. One rather consistent characteristic of CG models is their dependence on AA MD simulations. This has the negative consequence of including any undesirable characteristics of the AA model into the CG model. We have recently developed a novel parameterization approach that relies heavily on experimental data including surface tension, density and free energy and reduces the dependence on atomistic molecular dynamics simulations. The resulting CG potential is based upon rather standard functional forms facilitating implementation in conventional MD codes. This approach has been applied to nonionic and anionic surfactants, biologically relevent lipid molecules and amino acids. The results demonstrate the ability to make modular transferable CG sites that are capable of accurately predicting the phase and surface behavior specific to a system.
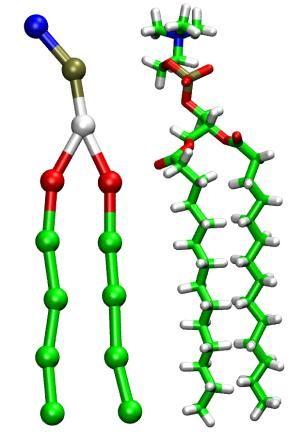
The image on the left shows the CG (left) and AA (right) representations of a DMPC lipid. The images on the right show the transition from a hexagonal phase to a lamellar phase of a PEG-lipid/water mixture upon reduction of the water content. Both are snapshots from simulations containing >1 mio. CG-beads.
CG models have been developed for non-ionic liquids which includes subsets of parameters for alkane, alcohol and ether based molecules. [1,2] Details of the water model can be found in this work as well. Our PC and PE based lipid model is available with flexibility in the hydrophobic chain length and degree of saturation. [3] Amino acids are currently in the development stage with some preliminary work reported. [4] We hope to bring the model up to speed soon. A very accurate phenyl based model with applications to fullerenes is available and has been integrated into our PC lipid model. [5,6,7] Ionic surfactants LABS [8] and SDS [9] along with counterions are also available. A more elaborate analysis of water models (not only including our current model) can be found in [10].
The specific functional forms required for the non-bonded potential of the CMM coarse grain model have been so far implemented into the following Molecular Dynamics simulation software packages:
LAMMPS (homepage)
LAMMPS is used by our group for most coarse grained simulations, and features excellent parallel scaling and many features. You have to enable the user-cg-cmm package (included in the main distribution) to use the CG potentials.
HOOMD-blue (homepage)
The HOOMD-blue package is MD program that runs entirely on graphics processors (GPUs) using the CUDA toolkit from Nvidia with up to 60x the speed of a single CPU.
MPDyn (homepage)
MPDyn is the "development" implementation of the CG model. Contact the MPDyn developer, Wataru Shinoda, to get access to a version that fully support the model. MPDyn is very efficient for serial execution and when using a small number of MPI tasks.
Tools to prepare and set up coarse grain simulations from scratch and from existing all-atom restart configuration are under development and will be published here, as well as the corresponding parameter and topology databased files.
References:
- W. Shinoda, R.H. DeVane, M.L. Klein, Mol. Sim. 33, 27-36 (2007)
- W. Shinoda, R.H. DeVane, M.L. Klein, Soft Matter 4, 2454 (2008)
- W. Shinoda, R.H. DeVane, M.L. Klein, J. Phys. Chem. B, 114, 6836 (2010)
- R.H. DeVane, W. Shinoda, P.B. Moore, M.L. Klein, J. Chem. Theor. Comput. 5, 2115-2124 (2009)
- R.H. DeVane, M.L. Klein, C. Chiu, S.O. Nielsen, W. Shinoda, P.B. Moore, J. Phys. Chem. B, 114, 6386 (2010)
- C. Chiu, R.H. DeVane, M.L. Klein, W. Shinoda, P.B. Moore, S.O. Nielsen, J. Phys. Chem. B, 114, 6394 (2010)
- R.H. DeVane, A. Jusufi, W. Shinoda, C. Chiu, S.O. Nielsen, P.B. Moore, M.L. Klein, J. Phys. Chem. B, 114, 16364 (2010)
- X. He, W. Shinoda, R.H. DeVane, K.L. Anderson, M.L. Klein, Chem. Phys. Lett. 487, 71-76 (2010)
- W. Shinoda, R.H. DeVane, M. L. Klein. In preparation. (2011)
- X. He, W. Shinoda, R.H. DeVane, M. L. Klein, Mol. Phys., 108, 2007-2020 (2010)
Collective Variables Module
Collective variables module main page
To tackle with the increasing complexity of macromolecular simulations, many techniques to enhance sampling have been made available in the past, most of them making use of reaction coordinates, or collective variables. Yet, multidimensional reaction pathways and conformational ensembles are most often modeled by very simple and ad hoc one-dimensional coordinates. Frequently this happens not because of methodological constraints, but rather because of too inflexible implementations within the MD programs used.
We have designed a module for MD codes that implements simultaneously several of such techniques, and an array of collective variables of broad usage. Currently available are: Adaptive Biasing Force (ABF), MetaDynamics (MtD), Steered MD (SMD) and Umbrella Sampling (US). There is no restriction on the number of variables, and many of their functional forms can be manipulated without recompilation. Rotational degrees of freedom are treated transparently, by generalizing the least-squares superposition approach of minimal RMSDs to model the orientations of macromolecules and flexible structures without artificial restraints. Statistical analyses can be performed at runtime, eliminating the need to store large trajectory files.
References:
- G. Fiorin, M. L. Klein and J. Hénin, Mol. Phys., in press (2013).
- J. Hénin, G. Fiorin, C. Chipot, and M. L. Klein, J. Chem. Theory Comput., 6, 35-47 (2010). (Link)
Contributed LAMMPS Packages
LAMMPS is a parallel molecular dynamics simulation software package with potentials for soft materials (biomolecules, polymers) and solid-state materials (metals, semiconductors) and coarse-grained or mesoscopic systems. It can be used to model atoms or, more generically, as a parallel particle simulator at the atomic, meso, or continuum scale. LAMMPS is designed to parallelize particularly well on large systems systems with no long-range interactions due to using a flexible spatial-decomposition of the simulation domain. The code provides a simulation framework that is easy to modify or extend with new functionality without losing parallel scaling.
Our contributions to LAMMPS include:
- fix imd for real-time visualization and interactive MD with VMD
- fix smd for steered MD
- pair_style cg/cmm etc. CMM coarse-grain pair potentials
- angle_style cg/cmm CMM coarse-grain angle/pair potential
Please see the Images and Movies Gallery page for some application examples.
Contributed VMD Plugins
VMD is a molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3-D graphics and built-in scripting. VMD supports computers running MacOS X, Unix, or Windows, is distributed free of charge, and includes source code. A member of the ICMS is a contributing VMD developer and also maintainting a number of VMD plugins listed below.
- TopoTools a Tcl script plugin library for manipulating topology information. It is meant to be a complementary tool to psfgen. TopoTools consists of a generic middleware script layer that provide convenient access to topology related data VMD. But it also has a number of high-level tools that allow reading and writing of topology file formats that cannot be parsed by molfile plugins, and replicating or combining multiple systems. TopoTools is bundled with VMD version 1.8.7 and later; updated versions may be found here.
- The signalproc plugin is a collection of scripted and compiled Tcl plugins that can be of use for signal processing and related types of applications within VMD.
- The gofrgui plugin provides a graphical user interface to measure gofr
- The irspecgui plugin is a GUI frontend to generating spectral densities.
- The clonerep plugin copies full sets of representations from one molecule to others.
- The dipwatch plugin allows to monitor the dipole moment of selections of atoms.
- The multimolanim allows to animate trajectories for systems with topology changes.
- CPMD trajectory file reader plugin
- X-Crysden xsf/axsf format reader
- HOOMD XML topology file reader.
Contributed HOOMD Modules
HOOMD-blue performs general purpose particle dynamics simulations on a single workstation, taking advantage of NVIDIA GPUs using the CUDA Toolkit to attain a level of performance equivalent a small cluster. Simulations are configured and run using simple python scripts, allowing complete control over the force field choice, integrator, all parameters, how many time steps are run, and more.
Our contributed modules to the HOOMD project for version 0.8.2 are:
