Membrane fusion is ubiquitous in biology: cell growth, extracellular transport and a multitude of processes involve membrane fusion. Fusion of synaptic vesicles to the plasma membrane is a key step in signal transmission between nerve cells. Within a cell, membrane budding and fusion enable non-disruptive transport of molecules between different organelles. Finally, viruses responsible for human disease, such as influenza and HIV, also begin the infection cycle by fusing their own membrane with the host cell's.
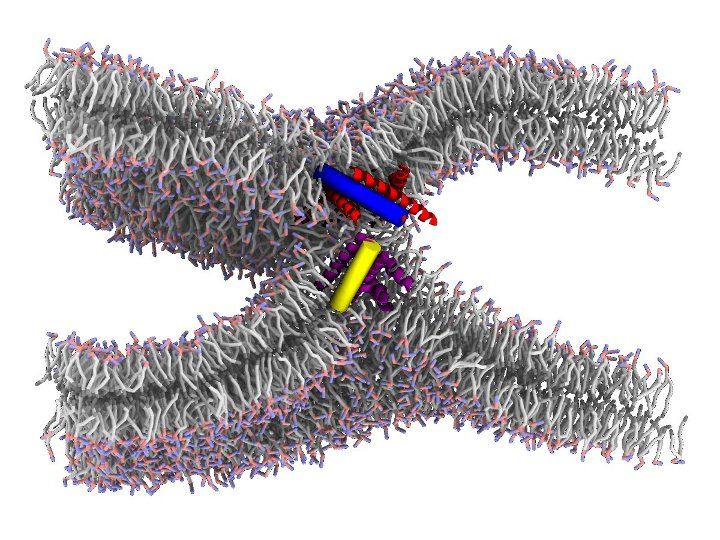
The physical mechanism of membrane fusion, wherein two lipid bilayers merge to form a continuous structure, is typically unfavorable and influenced by factors including lipid composition, hydration and electrostatics. However, in addition to lipids biological membranes contain embedded proteins, often with sizable extracellular footprints that can interact with other proteins or with the opposing membrane. Such fusion proteins are initially bound to one of the two fusing membranes via a transmembrane helix domain, and utilize energetically favorable conformational transitions to lower the activation energy for membrane fusion.
It is well understood that fusion proteins shape the initial stages of the process, by facilitating the apposition between bilayers, and their dehydration as they come into more intimate contact. However, a major open question concerns the specific roles played by fusion proteins in the later stages of fusion. The “real” situation is likely to be intermediate between two hypothetical extreme scenarios: in the “lipid-centric” scenario, fusion proteins hold bilayers in close proximity, but remaining outside the point of membrane apposition, which is instead made up exclusively of lipids; in the “protein-centric” scenario, fusion proceeds via a pore lined with proteins, which remain at the very center of the point of contact as their conformational changes progress toward more stable states.
We are using simulations, all-atoms and coarse grained, combined with advanced sampling techniques, to test both scenarios on realistic examples of fusion. Our goal is to obtain a predictive computational model, to be used in understanding fundamental processes such as the transport of neurotransmitters, the design of new delivery systems and the infection by viruses.
References:
- J. E. Donald, Y. Zhang, G. Fiorin, V. Carnevale, D. R. Slochower, F. Gai, M. L. Klein, W. F. DeGrado Proc. Natl. Acad. Sci., 108(10), 3958-3963 (2011) (Link)
Translocation of Carbon Nanospheres Through Lipid Membranes
Carbon nanospheres (CNS) can be used for pharmaceutical and biomedical applications, such as drug delivery or cell imaging. It has been observed that such objects can enter into the nucleus of cells without any specific surface active chemical molecules. On the other hand there is the question whether such particles are toxic for human cells.

The translocation of carbon nanospheres through cell membranes is not fully understood on molecular scale, specifically nano-objects of sizes that are close or larger than the thickness of typical lipid membranes (around 3nm). In this study we investigate the mechanism of translocation of moderatly hydrophobic compounds through membranes without attaching any functional molecules on the translocated molecule. Fullerenes of different sizes serve as model system. We are specifically interested in the free energy change when a CNS enters the membrane.
We use all-atom Molecular Dynamics (MD) simulations to address this question. For CNS with sizes exceeding the bilayer thickness we develop a coarse grain model. Coarse grain simulations help to to overcome the required length and time scales, allowing for example to study also structures of CNS inside the membranes. The image shows a simulation snapshot of a C540 fullerene at the lipid-water interface.
References:
- A. Jusufi, R. H. DeVane, W. Shinoda, M. L. Klein, Soft Matter, 7, 1139 (2011). (Link)
